FISH Probes
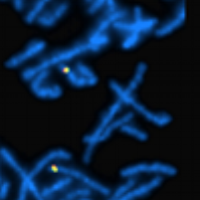
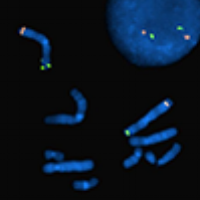
Fluorescence in situ hybridisation (FISH) is a molecular diagnostic technique utilising labelled DNA probes to detect or confirm gene or chromosome abnormalities. It is often used in cancer diagnosis. The sample DNA (metaphase chromosomes or interphase nuclei) is first denatured, a fluorescently labelled probe of interest is then added to the denatured sample mixture and hybridises with the sample DNA at the target site as it re-anneals back into a double stranded DNA. The probe signal can then be seen through a fluorescent microscope and the sample DNA can be scored for the presence or absence of the signal. Unlike most other techniques used to study chromosomes, FISH does not have to be performed on cells that are actively dividing. This makes it a very versatile procedure. Uses encompass a wide range of applications such as the detection of aneuploidy, constitutional microdeletion syndromes as well as rearrangements. These aberrations have clinical implications for numerous genetic diseases such as leukaemia, lymphoma, solid tumours, autism and other developmental syndromes. FISH probes are commonly made from BAC clones. Pisces Scientific in partnership with Empire Genomics provides access to over one million BAC clones from the Roswell Park Cancer Institute genomic libraries, which are available for purchase as either unlabelled or custom labelled FISH probes.
Custom FISH Probes
Empire Genomics specialises in custom development of FISH probes based on customer specifications. Our custom FISH solutions extend beyond single gene, single region analysis. Our FISH probe panels utilise multiple probes that are each labelled with a unique fluorophore. This allows for the simultaneous identification of several possible anomalies in a single sample. Empire Genomics will work with you to design custom FISH probe panel(s) for simultaneous coverage of larger regions of interest, which may require the design a probe panel for full coverage of specific diseases. All Empire Genomics FISH probes are directly labelled using standard Nick Translation protocols and are provided with hybridisation buffer solution.
Gene specific probes
Empire Genomics manufactures locus-specific probes that are ideal for the rapid identification of a range of chromosomal aberrations across the genome. Specific to regions of interest in oncology and constitutional syndromes, our catalogue probes are designed for the purpose of detecting and defining various rearrangements, deletions and trisomies frequently observed in these areas.
chromosome control probes
Standard controls are typically utilised in cytogenetic laboratory testing, including FISH studies. For metaphase FISH, it is recommended that FISH tests include control probes to tag the chromosome(s) of interest. Such probes afford a limited level of quality control by providing an internal control of hybridisation efficiency. The target sequence on a normal chromosome serves as the best control of technical variables. If a probe is used that does not have an inherent chromosome control signal (ie, an X or Y chromosome probe analysis in a male with clonal loss of the Y chromosome), another sample that is known to have the probe target (a normal 46, XY male for this example) should be run in parallel with the patient sample.